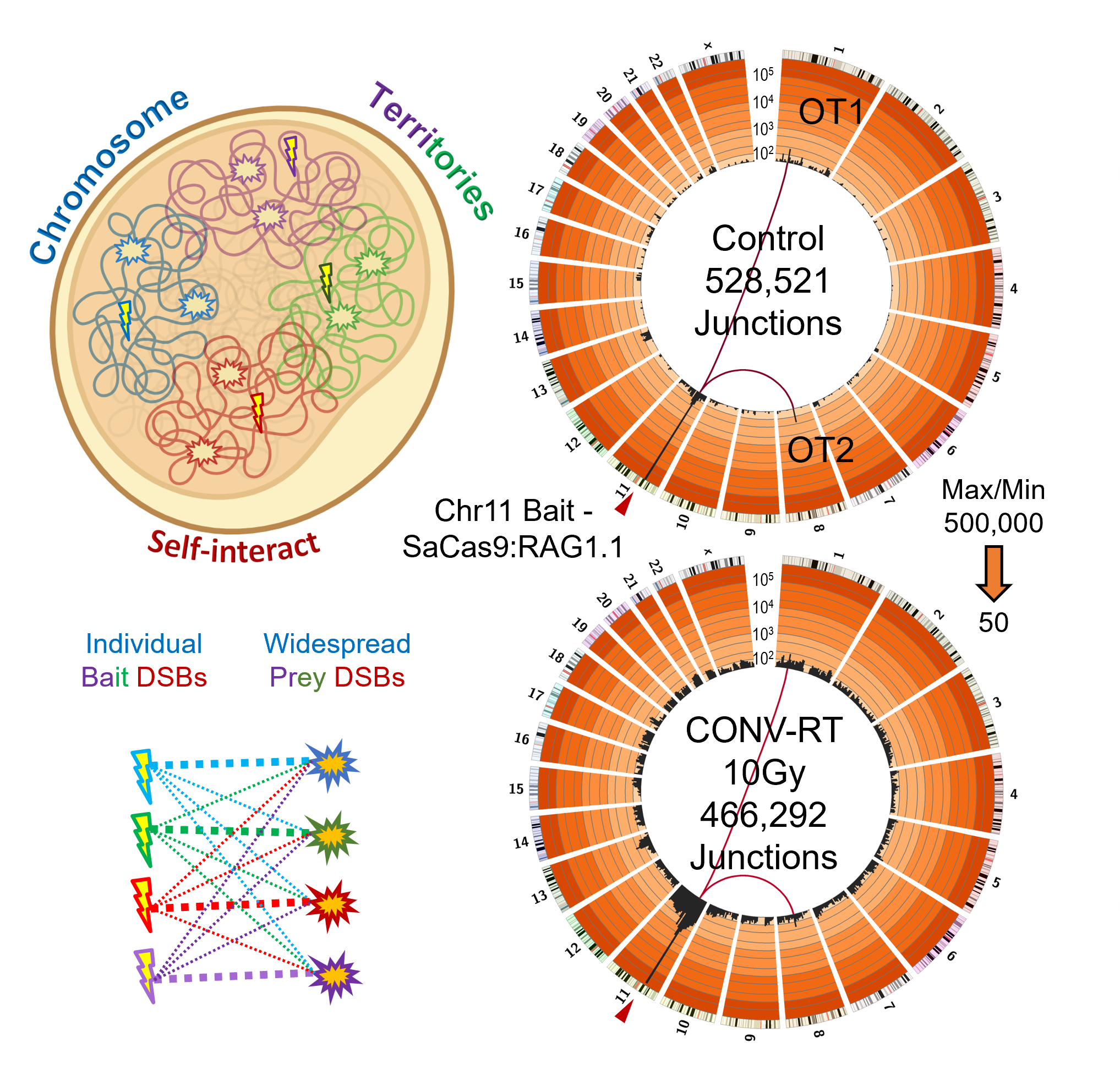
Frequent DNA:DNA contacts of DSB sites increase odds to translocate
Low-level and distributed genome-wide translocations to bait DSBs emphasize their relative DNA interaction frequencies. Specifically, they indicate a measurable level of genome instability and reveal aspects of genome organization hierarchy. This can be viewed by modulating the level of available DNA ends generated (i.e. ionizing radiation, oxidative free radicals, aberrant nuclease activity, topoisomerase adducts, replication fork stalling, etc.)
Widespread DSBs make the chromosome harboring the bait DSB a hotspot for translocation
Conventional Radiotherapy (CONV-RT) from an electron LINAC generates widespread DSBs, but low level at any given location, whether their translocation frequency represents the DNA:DNA contact frequency near a bait DSB. The greater local translocation competition decreases the frequency of recurrent translocations, for instance, to off-target (OT) sites.
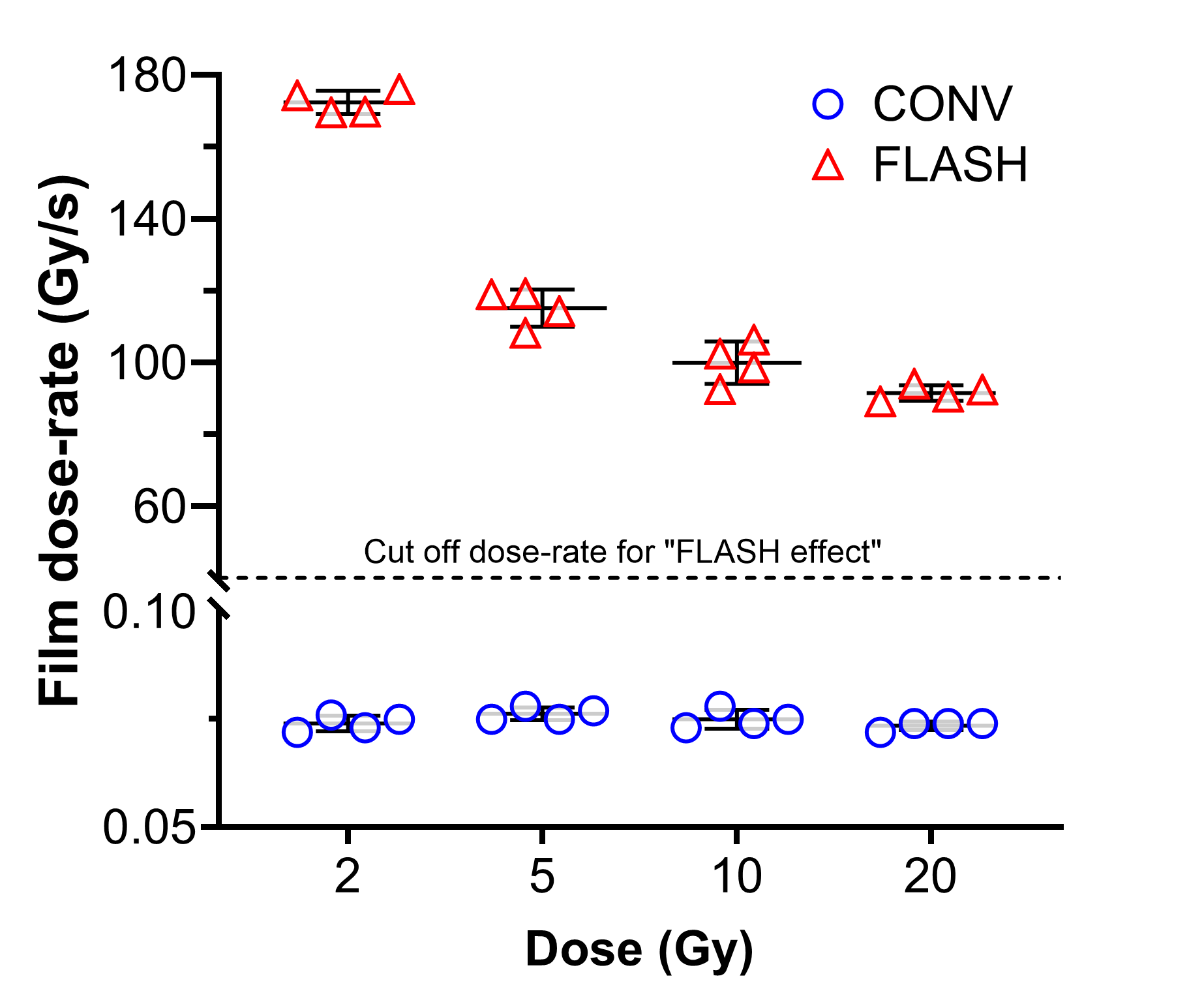
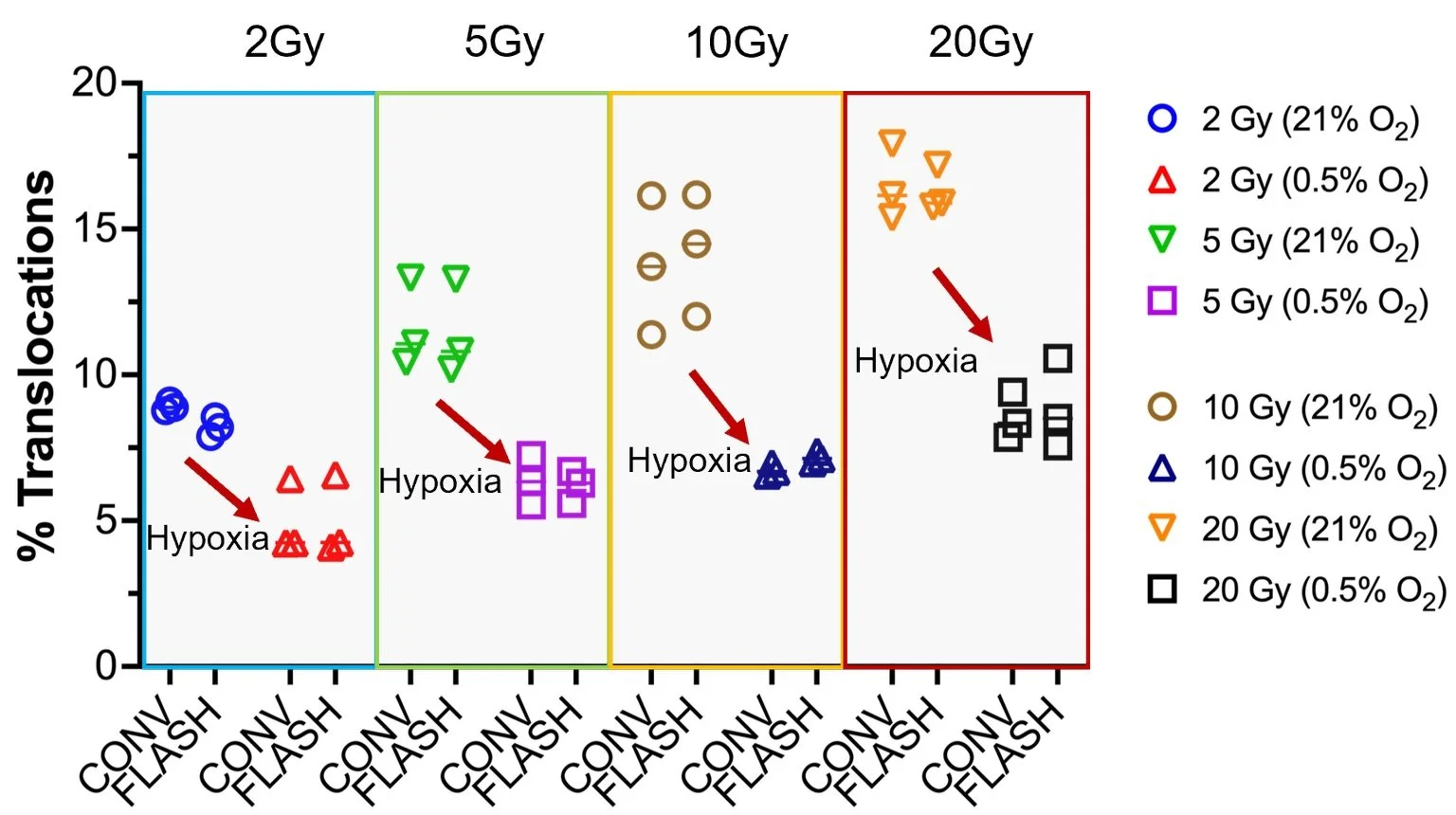
High dose-rates of ionizing radiation (>40Gy/sec; FLASH) facilitate normal tissue sparing while retaining high level tumor control. While the mechanism of FLASH-IR is not known, translocation differences at low or high oxygen tensions are not apparent across a range of IR doses. See Barghouth et al., 2023 for more details.