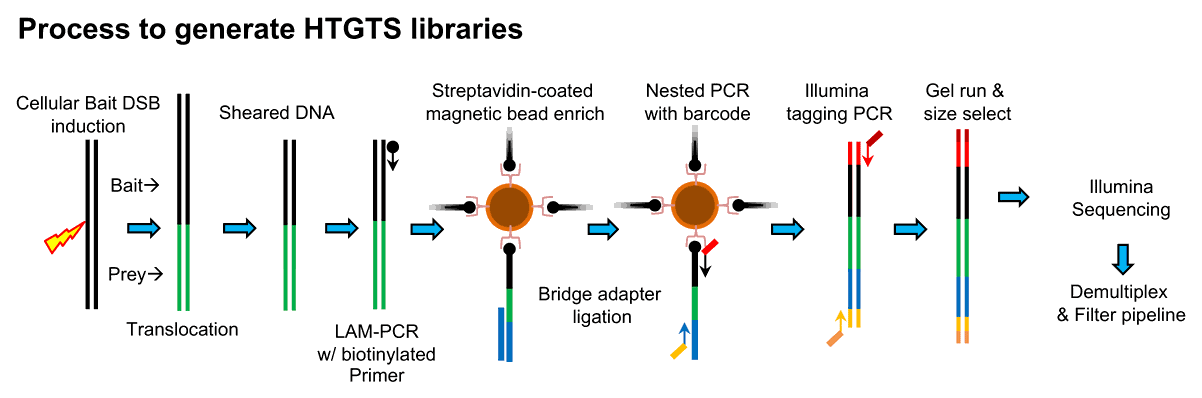
The LAM-HTGTS platform
High-throughput Genome-wide Translocation Sequencing (HTGTS) was developed originally to identify recurrent “prey” DSBs genome-wide using minimal selective forces (Chiarle et al., 2011). A second generation of the assay was developed, involving linear amplification (LAM) with a single primer, ligation of the single DNA strand using thermodynamically stabilizing bridge adapters (LAM-HTGTS) (Frock et al., 2015; Hu et al., 2016) and a complete overhaul of the junction analysis pipeline to enable single nucleotide resolution measures of translocation bait/prey junctions. LAM-HTGTS has served as the basis for several downstream applications including antigen receptor repertoires (Lin et al., 2016), DNA:DNA interactions (Jain et al., 2018), and comprehensive end-joining measures in non-dividing cells (Liang et al., 2021).
A long-term focus is to further develop/adapt this platform for basic research and translational studies
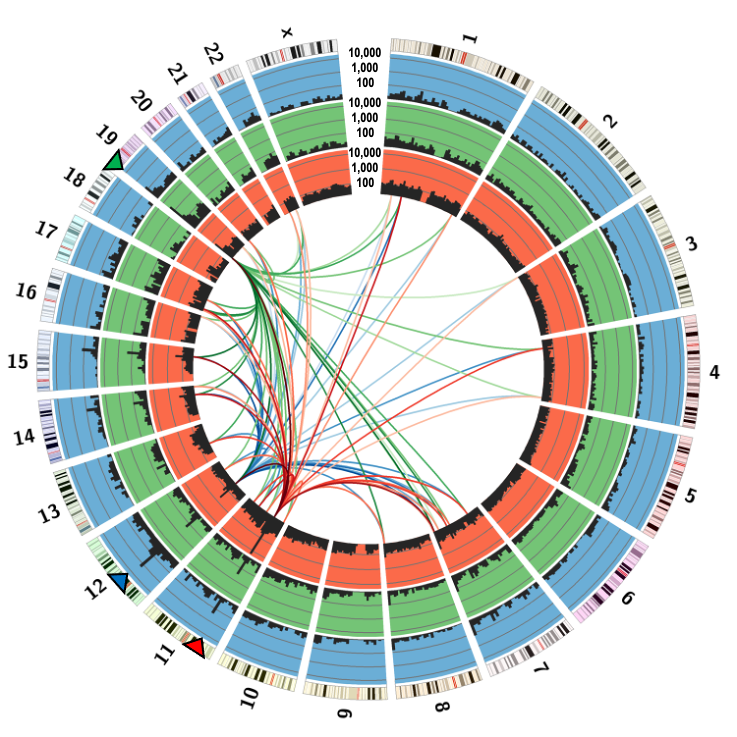
Key feature: Readily detectable frequent DSBs
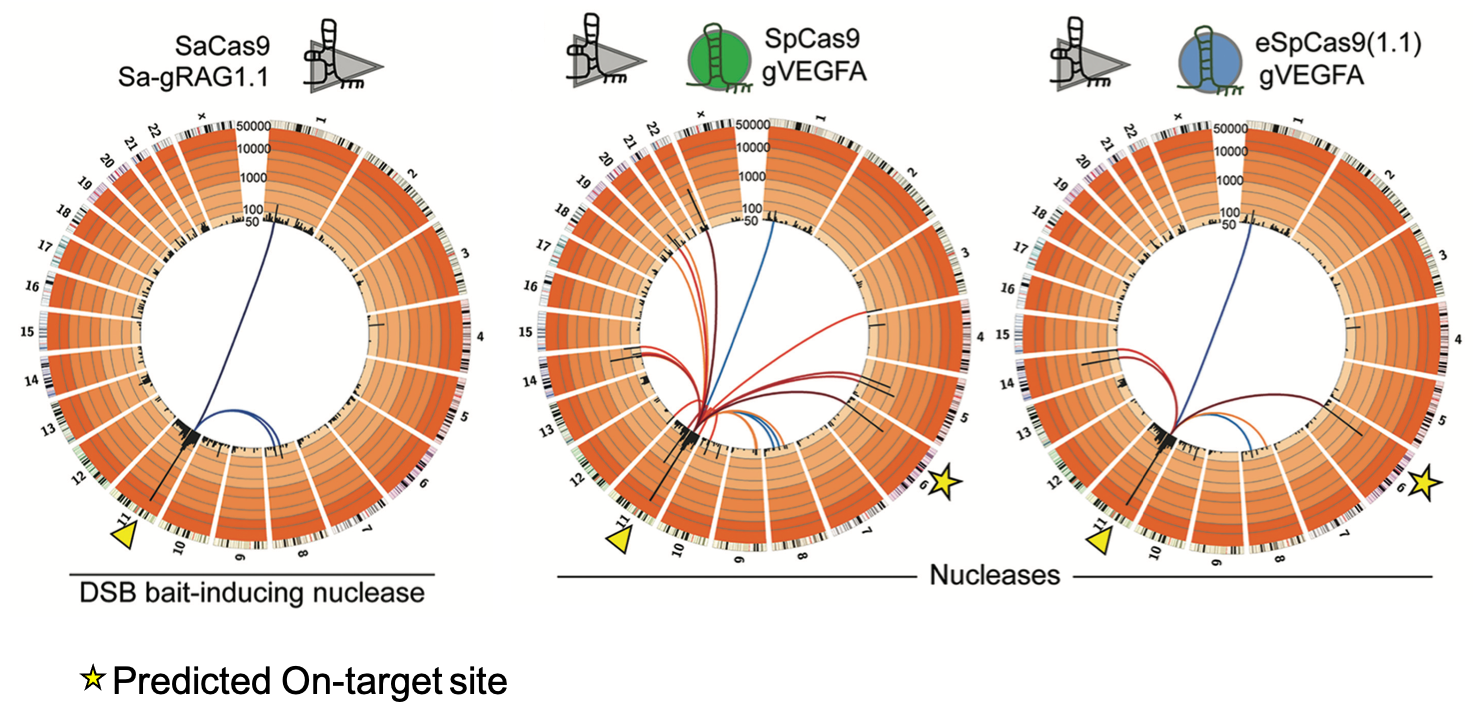
Universal bait DSB approach to evaluate endonuclease specificity & efficacy
Orthogonal HTGTS: using different species Cas9 bait / prey DSBs (S.aureus - Sa / S.pyogenes - Sp) to compare engineered SpCas9 variant, eSpCas9(1.1). Collaboration with the Gonçalves group at Leiden University (Wang et al., 2021; Chen et al., 2020).
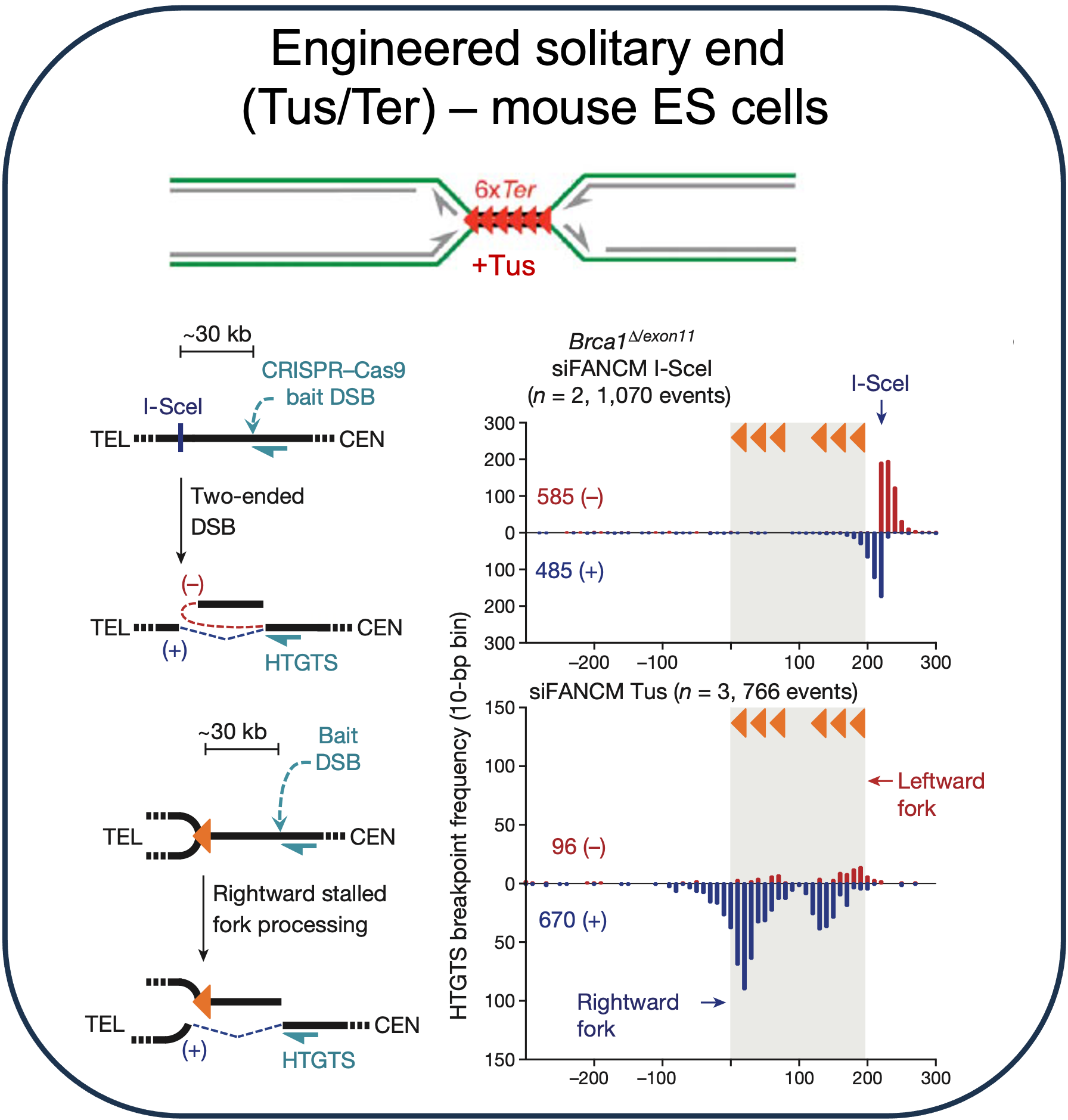
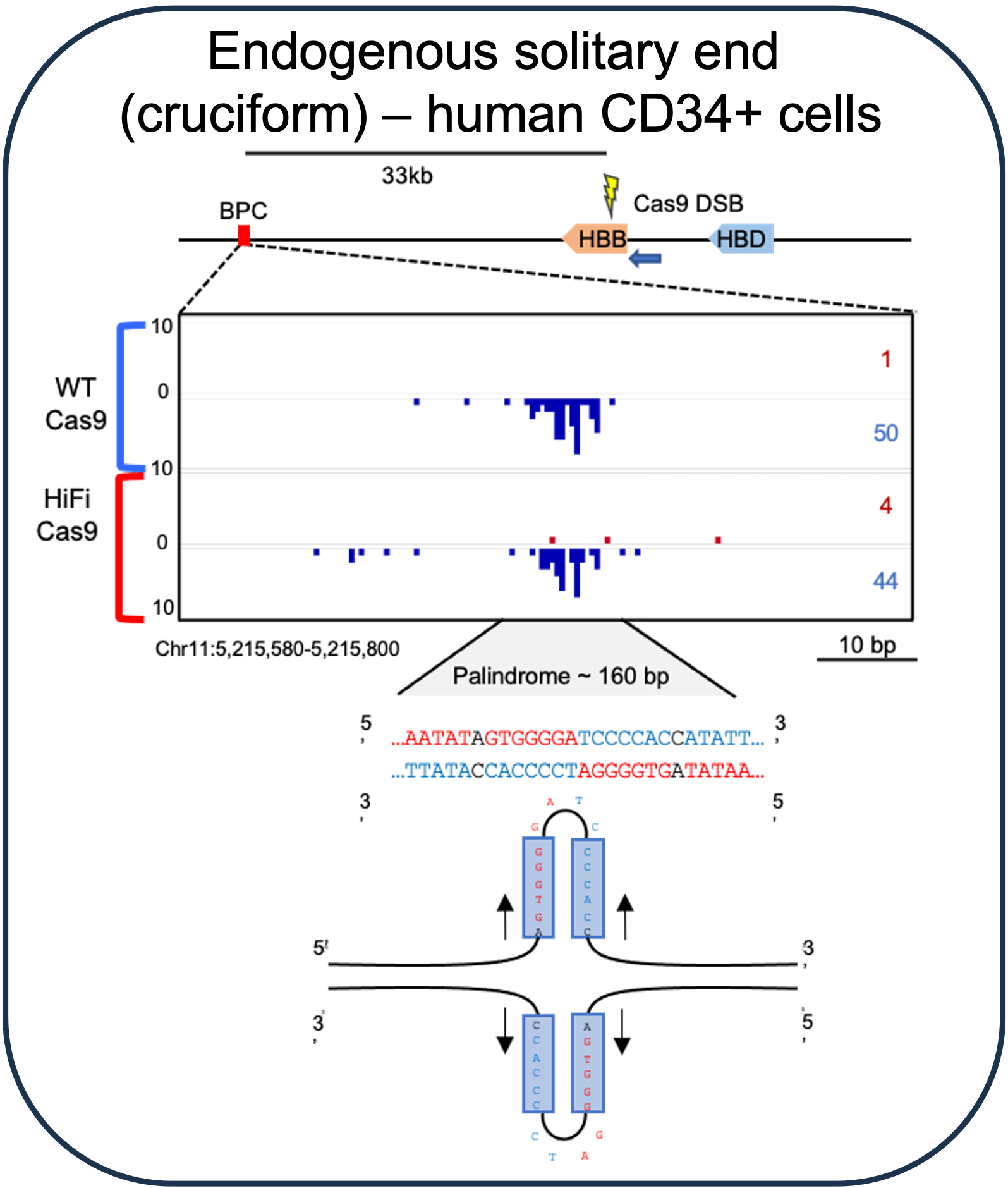
Recurrent solitary ends of replication-stalled forks
Solitary ends from stalled replication forks are substrates for translocation (note strand orientation bias) and are readily apparent at engineered (Willis et al., 2017) or endogenous (Lattanzi et al., 2021) locations where DNA strand separation is more difficult.