reJoin & Translocation sequencing
(HTGTS-JoinT-seq)
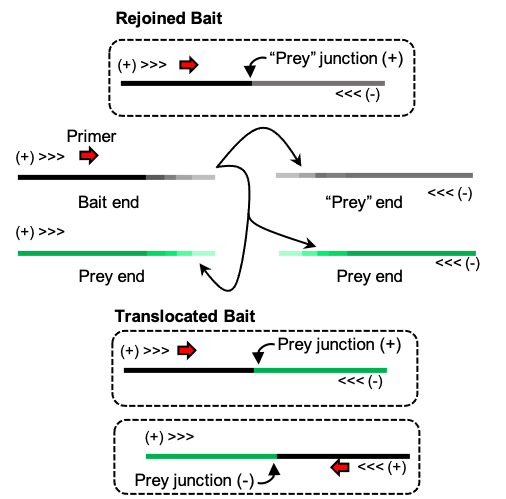
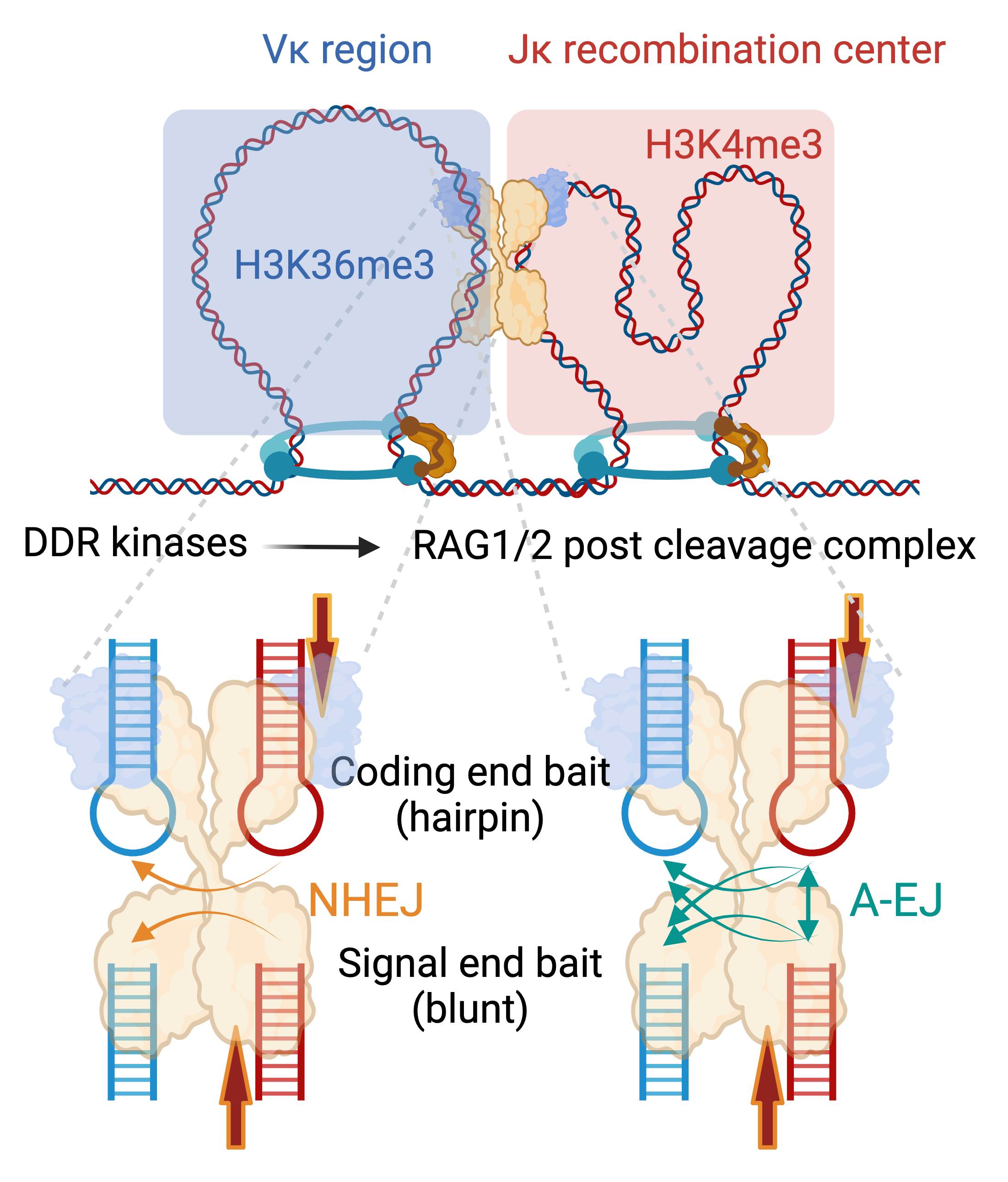
Translocations are rare DNA repair events. For such events to occur, the DSB rejoining process must be disrupted such that DNA ends can still synapse but with different partners. Thus, understanding end (re)synapsis is a crucial step towards developing tools to modulate translocation rates. We developed JoinT-seq to distinguish between such events as they represent different stages of DSB repair and could differentially implicate repair pathways, guided by the DNA Damage Response.
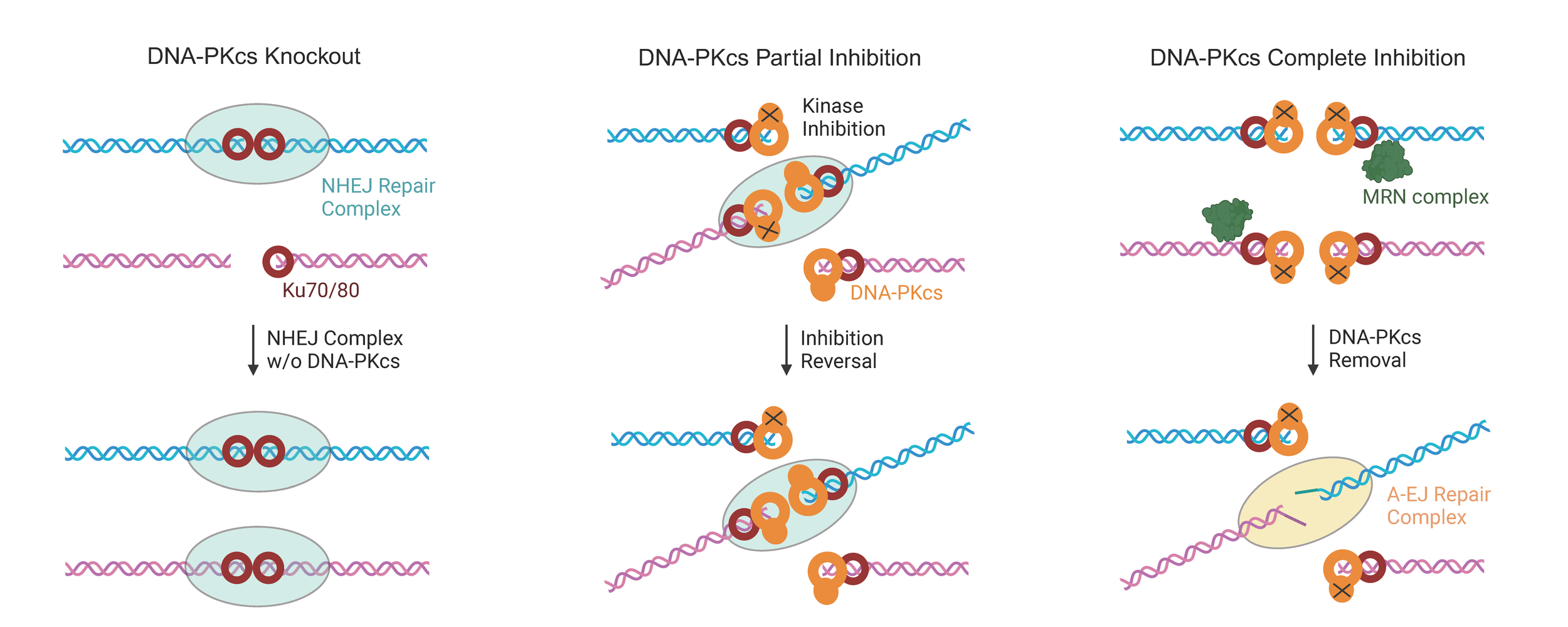
Disrupting the ratio between DSB rejoin versus translocation could be pathway intrinsic or involve transitioning to another pathway. Check out our recent work on DNA-PK kinase inhibition impacting translocations and repair pathway choice (Wang et al., 2024a)
Distinct Alternative end-joining (A-EJ) mechanisms in G0/G1 phase
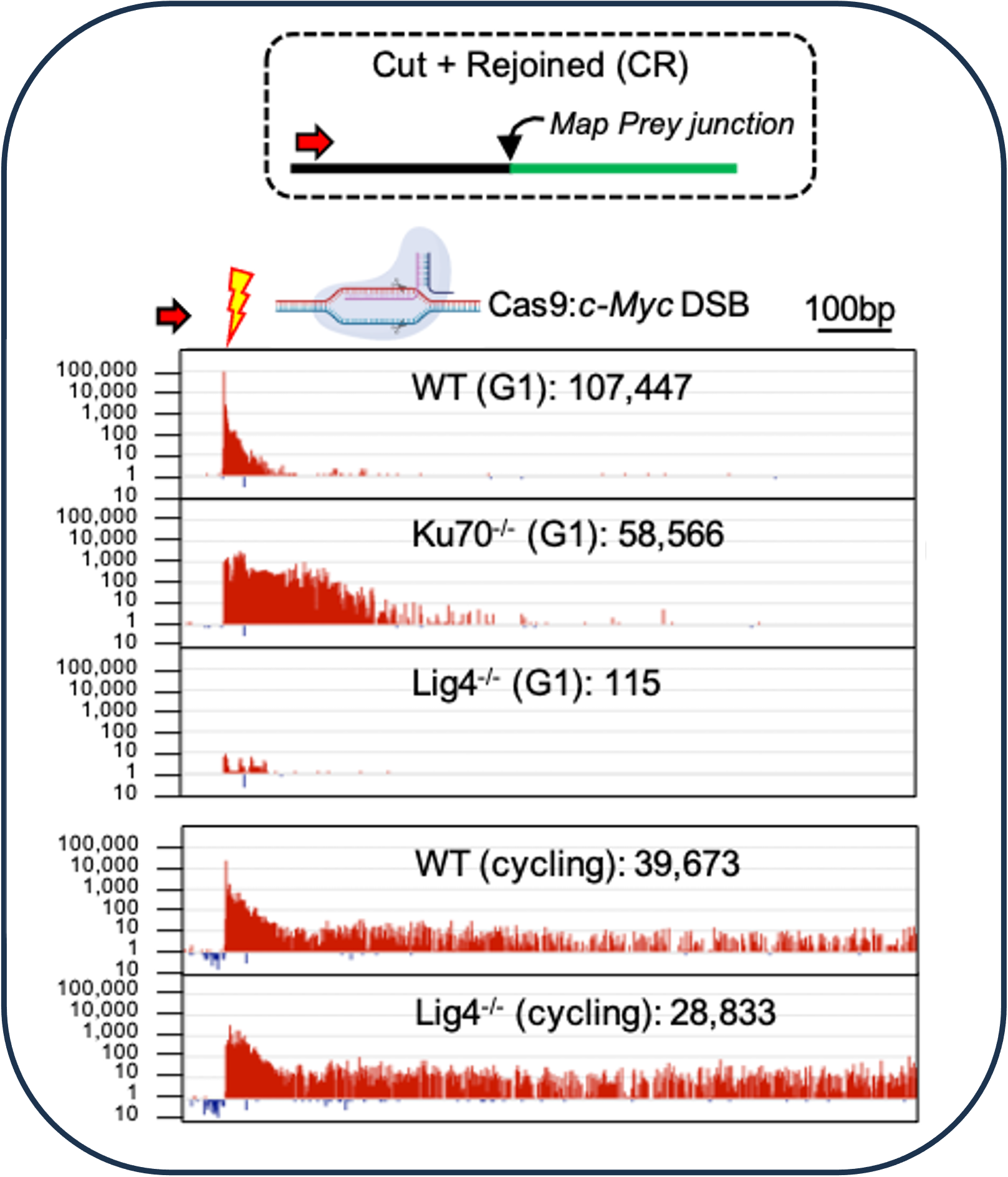
Limited alternative end-joining in the absence of Lig4 but only in G0/G1 phase (Liang et al., 2021)
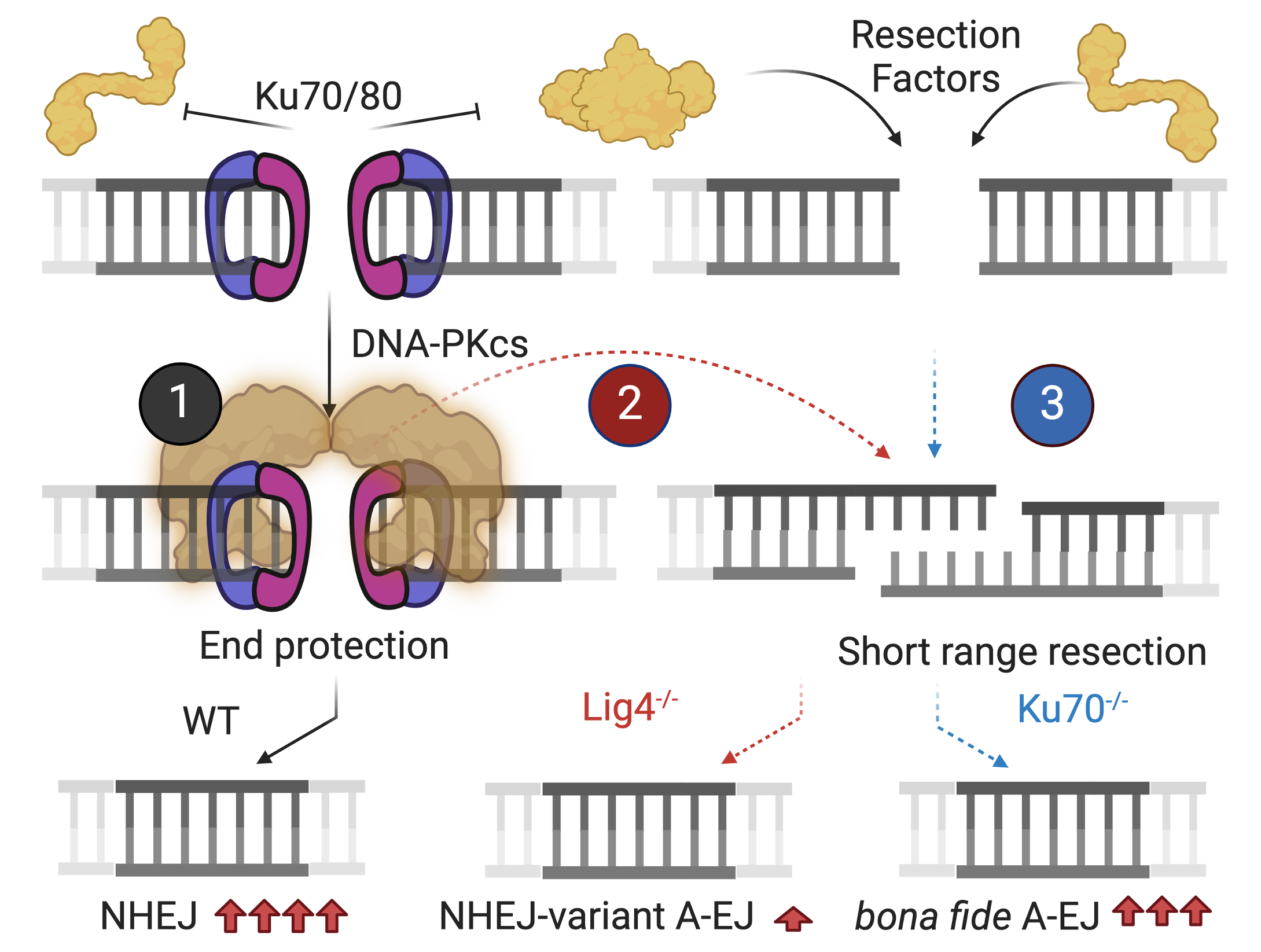
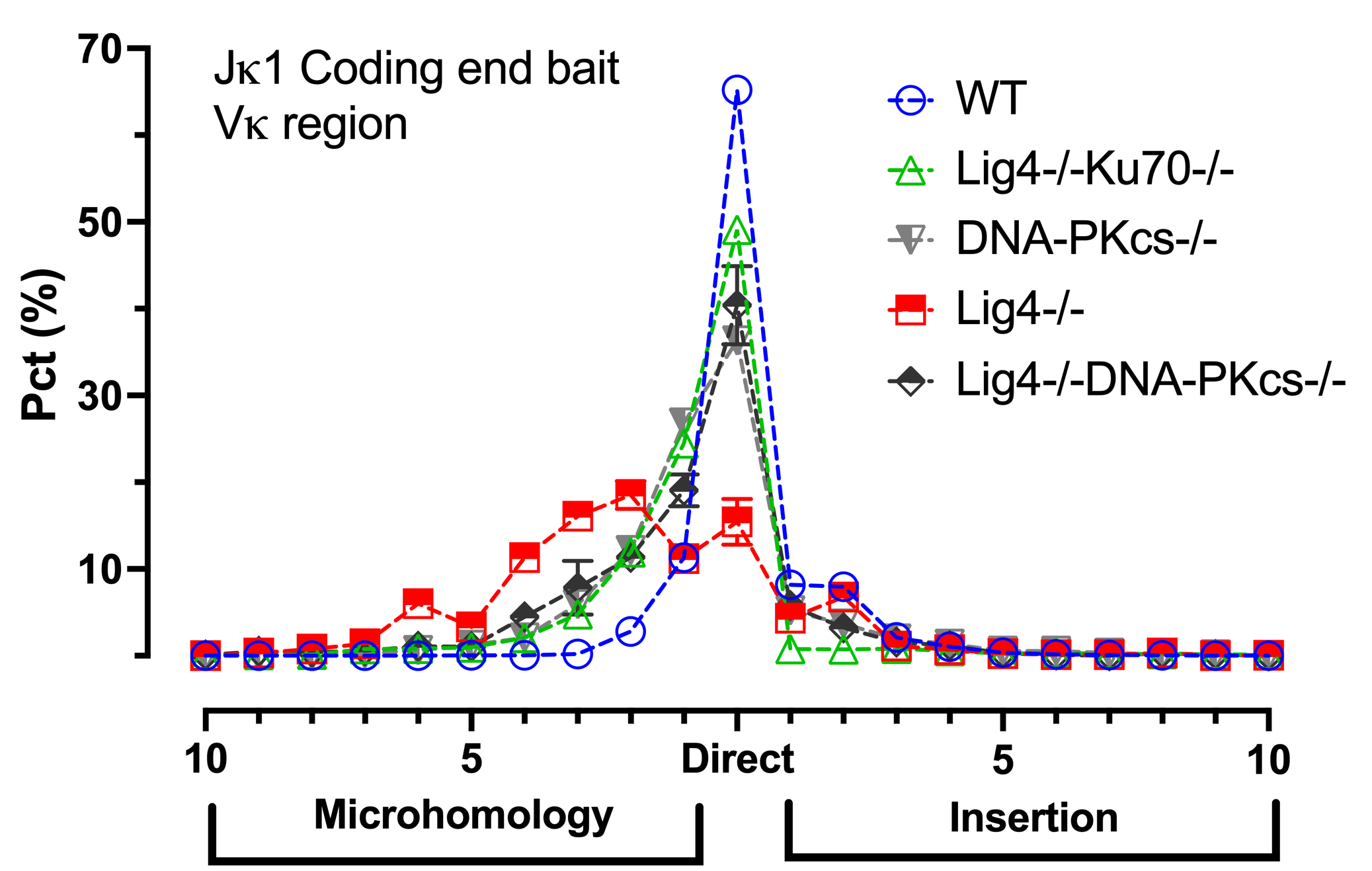
Nonhomologous end-joining (NHEJ) core components include the Ku70/80 (Ku) DSB sensor and the XRCC4/Lig4 ligation complex. Under G0/G1 arrest, the absence of Ku reveals a robust bona fide A-EJ pathway while the absence of XRCC4/Lig4 reveals an NHEJ-variant A-EJ pathway that is more active in S-G2/M cell cycle phases.
In the context of coding end V(D)J recombination, using the Ig kappa (Igk) locus, NHEJ and the two A-EJ pathways (NHEJ-variant and bona fide) are readily discerned at the level of microhomology utilization at recombined Vk-Jk joints (Wang et al., 2024a)
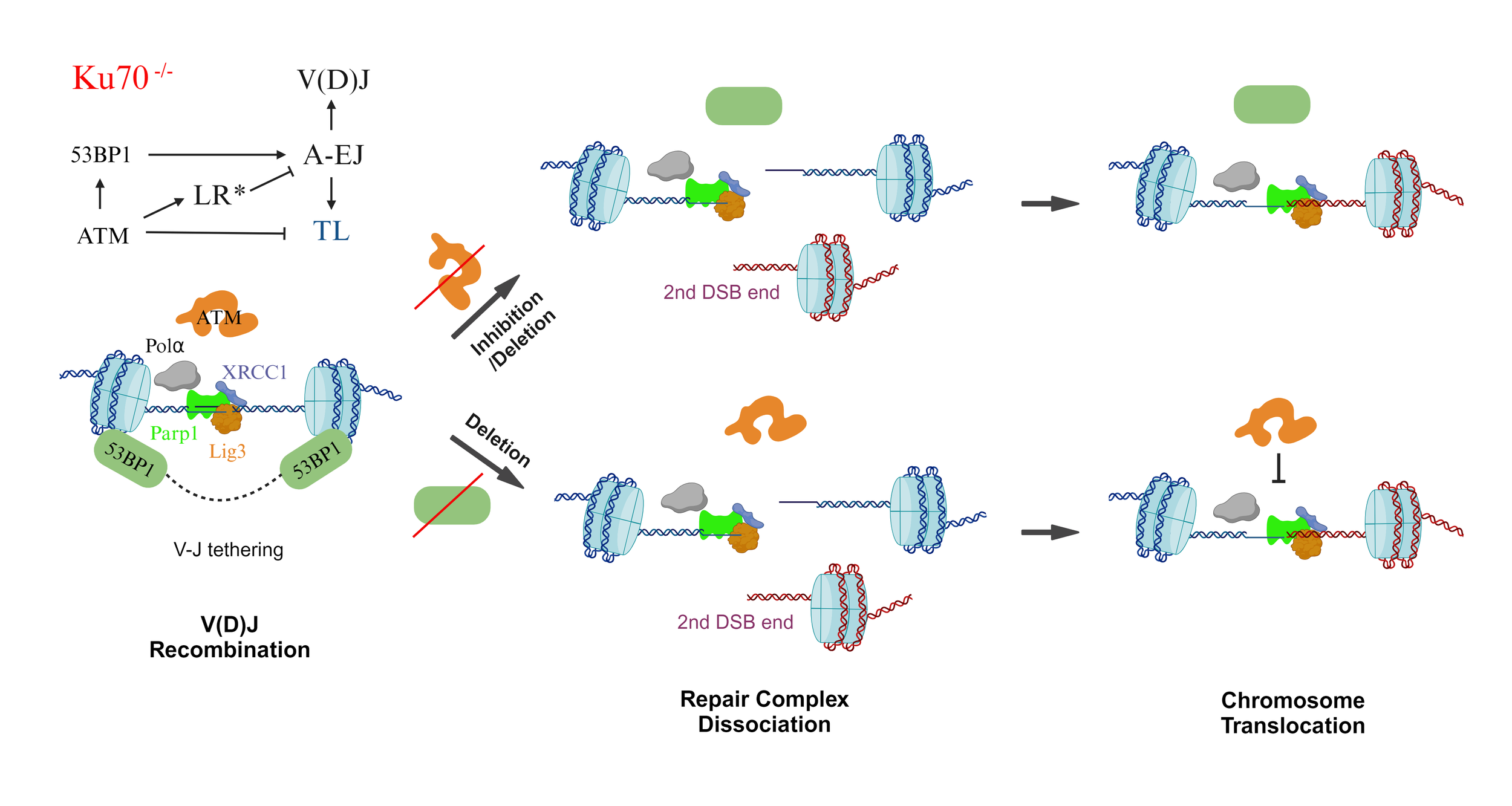
What is the bona fide A-EJ pathway?
Components of Base Excision Repair collaborate with the ATM-initiated DDR to suppress MMEJ. XRCC1 and 53BP1 are central in facilitating core factor functions and overall repair capacity, respectively. Parp1 and ATM together robustly suppress MMEJ, like XRCC1 alone. We posit that most MMEJ activities likely occur beyond the 53BP1 chromatin domain, where long-range resection (LR) suppresses the bona fide A-EJ pathway (Wang et al., 2024b).